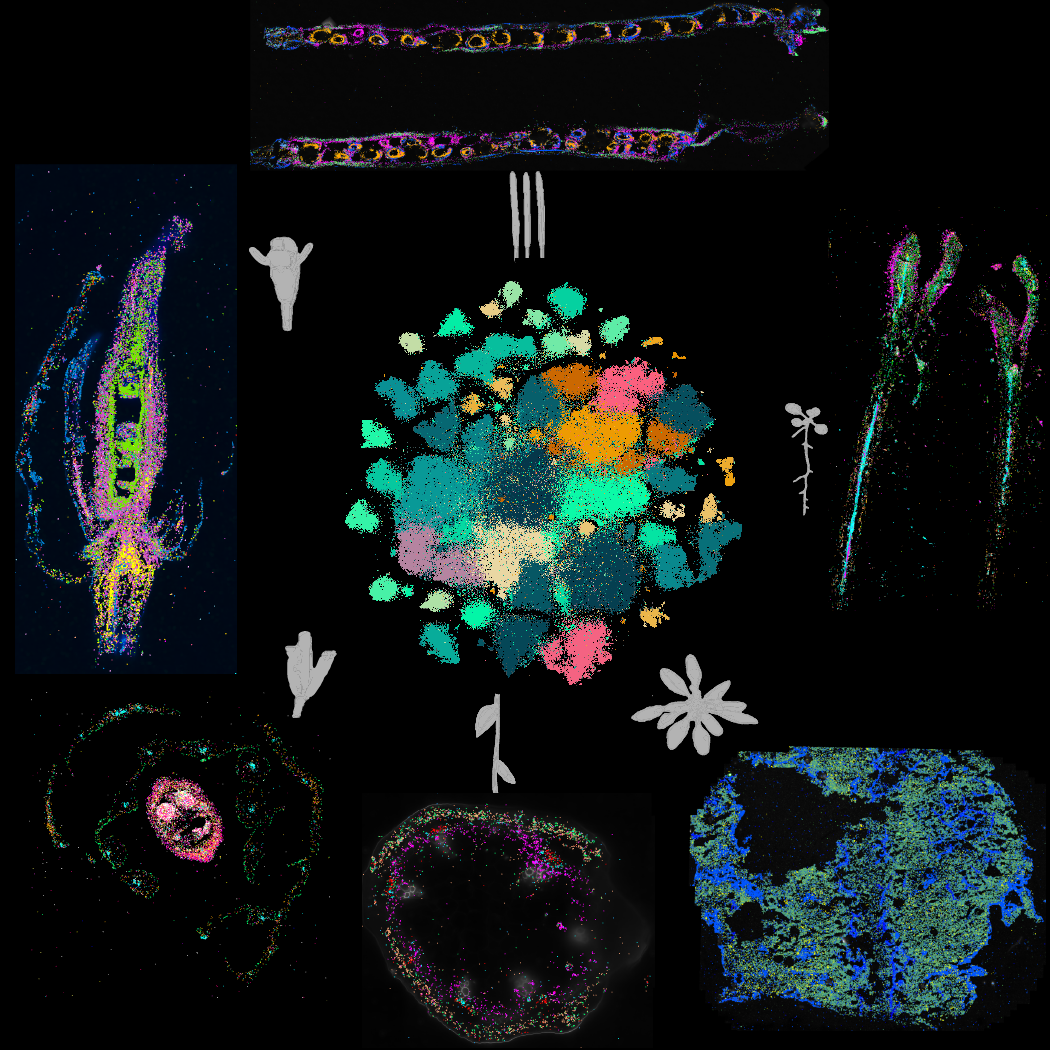
Incredible tissue diversity exists throughout Arabidopsis development; from seeds to, roots, leaves, stems, and reproductive tissues, and the composition and arrangement of cells and cell types within. To survey cell type diversity throughout development, we applied single-nucleus RNA sequencing to whole tissues and organs at ten distinct developmental timepoints, resulting in over 400,000 nuclei that represent 183 clusters.
This browser enables the exploration of each of our single nucleus transcriptome datasets, and may serve as a reference for cell type specific and developmental specific expression of genes throughout the entirety of Arabidopsis lifecycle.
For a brief tutorial on data exploration click here
Please Note: when accessing larger datasets, it may take several minutes to load the dataset

Access the preprint at:

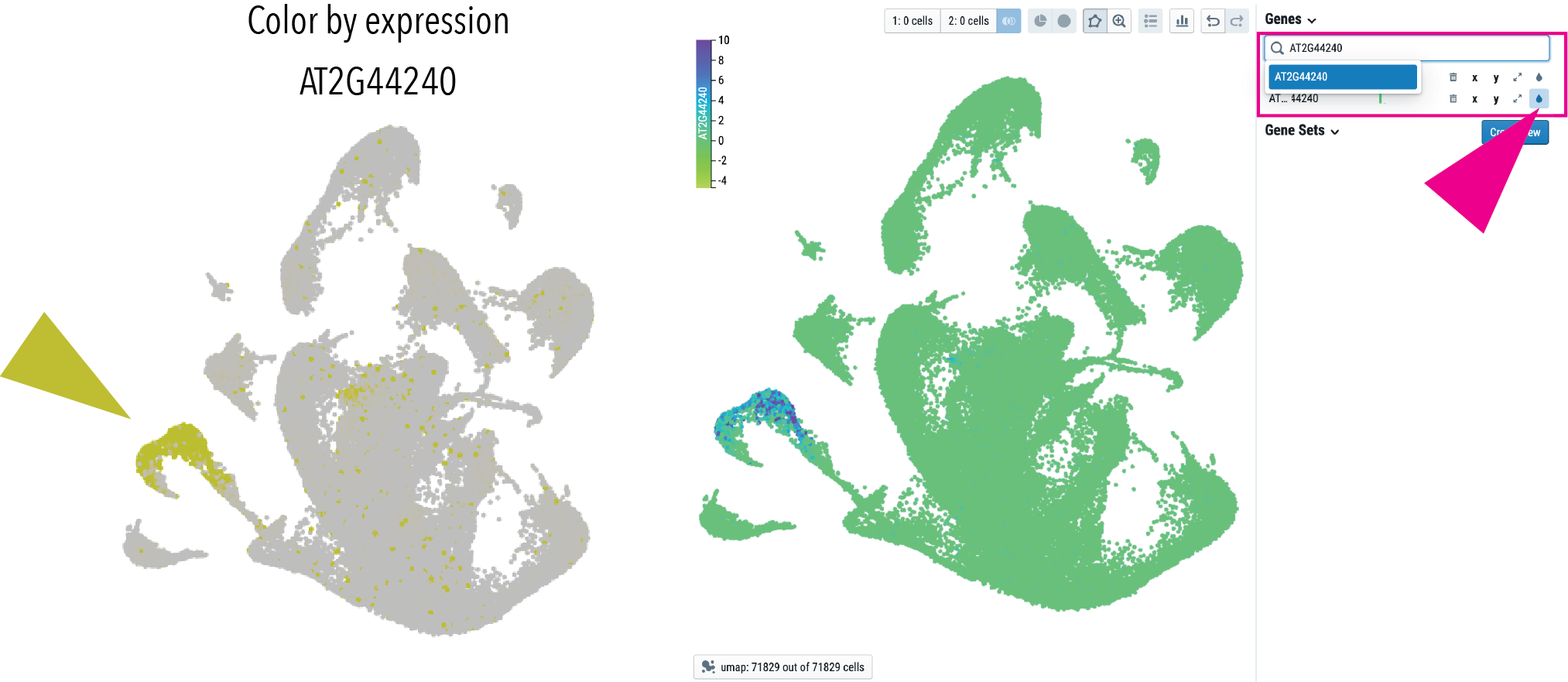
Input your gene of interest into the field and click button indicated to color by your gene of interest. Scaled expression values of the all transcripts are displayed.
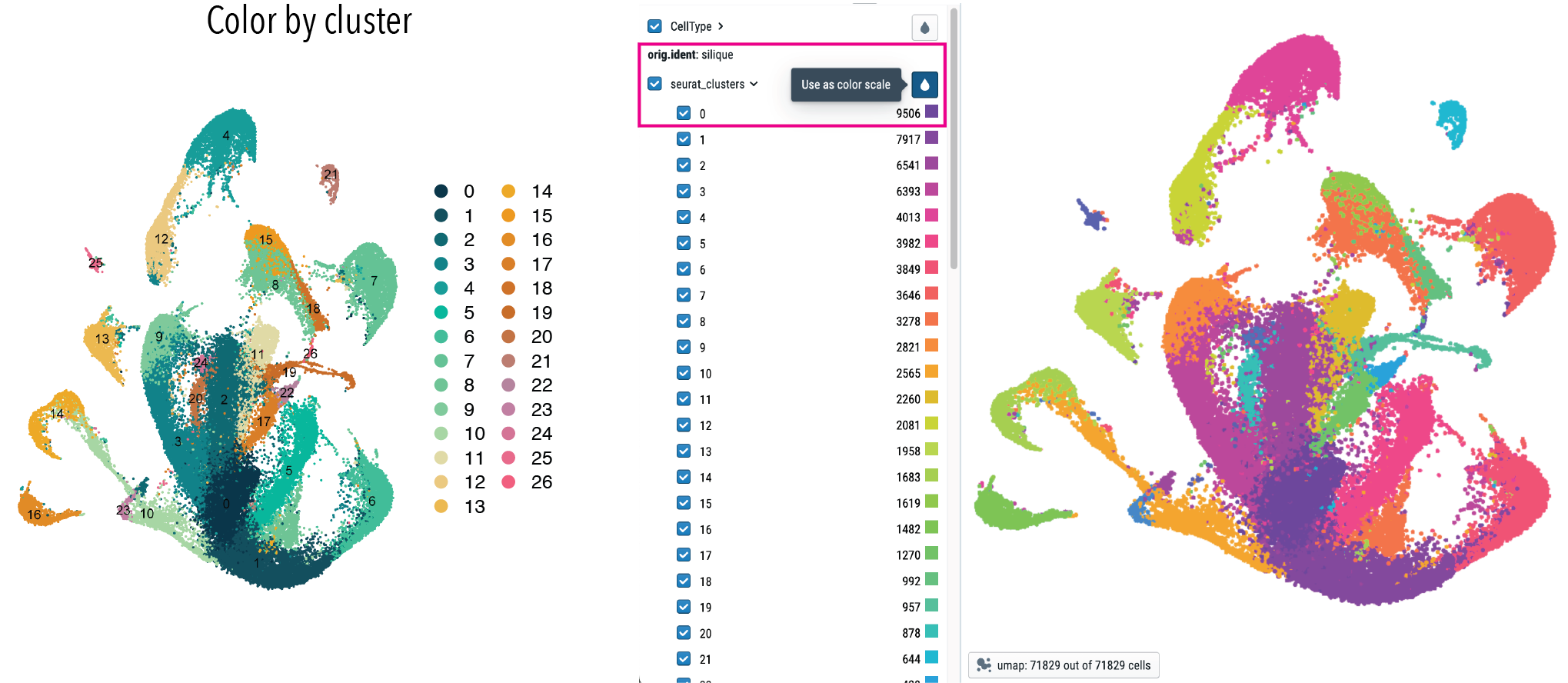
Click here to color the graph by cluster number
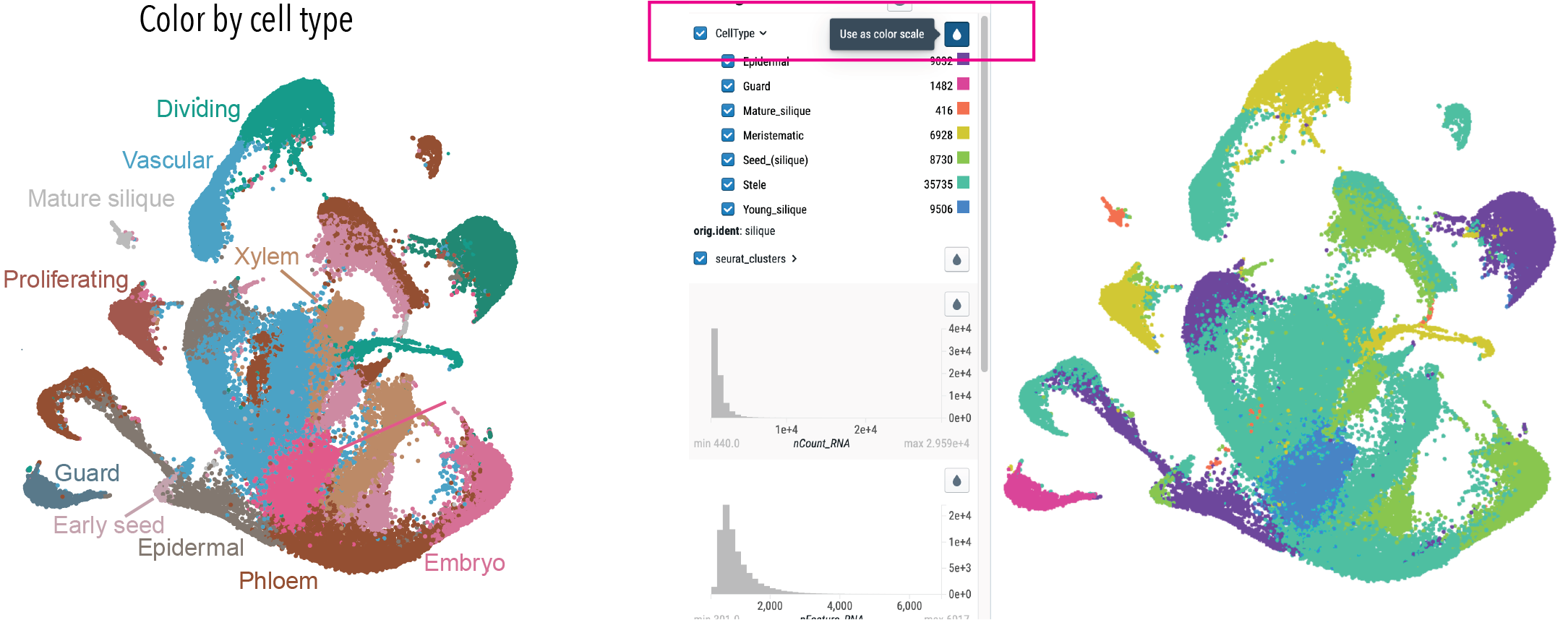
Click here to color the graph by cell type annotation
For more in-depth CELLxGENE resources visit CELLxGENE
Register and download the Vizgen MERSCOPE Visualizer Software

Guided tutorial for the Vizgen MERSCOPE Visualizer Software coming soon
Raw sequencing data and processed data are available at GEO accession: GSE226097
All processed MERFISH spatial datasets can be found at the following link
A link to download the spatial imaging data can be found here
To interact with the processed spatial data download the Vizgen Visualizer software
Interactive data browsing utilizes CELLxGENE v1.1.1
Dataset serving utilizes CELLxGENE-gateway v3.0.10
Scripts used to analyze the datasets can be found on GitHub (coming soon)
The Arabidopsis developmental atlas is maintained by Travis Lee
May 2025
Single-cell datasets of three flower spatial MERFISH datasets are now available
April 2025
Cluster annotation of the 3d-old-seedling, stem, and flower datasets has been updated based on spatial transcriptomic validation
January 2025
Processed MERFISH spatial datasets of seedling, stem, and flower datasets can now be found at the following link
Additional updates will be arriving soon
August 2023
Now browse the normalized expression of all transcripts detected within each dataset.
July 2023
Find the processed silique MERFISH dataset here
To interact with the processed data download the Vizgen Visualizer software
March 2023
We are live! Interact and browse the expression of up to 2,000 of the highly variable genes identified in each of the 11 individual datasets and the integrated rosette and seedling time course datasets.